Stabilised mosaic single cell data integration using unshared features
Source:R/stabMap.R
StabMap.RdstabMap performs mosaic data integration by first building a mosaic data topology, and for each reference dataset, traverses the topology to project and predict data onto a common principal component (PC) or linear discriminant (LD) embedding.
stabMap(
assay_list,
labels_list = NULL,
reference_list = sapply(names(assay_list), function(x) TRUE, simplify = FALSE),
reference_features_list = lapply(assay_list, rownames),
reference_scores_list = NULL,
ncomponentsReference = 50,
ncomponentsSubset = 50,
suppressMessages = TRUE,
projectAll = FALSE,
restrictFeatures = FALSE,
maxFeatures = 1000,
plot = TRUE,
scale.center = TRUE,
scale.scale = TRUE
)Arguments
- assay_list
A list of data matrices with rownames (features) specified.
- labels_list
(optional) named list containing cell labels
- reference_list
Named list containing logical values whether the data matrix should be considered as a reference dataset, alternatively a character vector containing the names of the reference data matrices.
- reference_features_list
List of features to consider as reference data (default is all available features).
- reference_scores_list
Named list of reference scores (default NULL). If provided, matrix of cells (rows with rownames given) and dimensions (columns with colnames given) are used as the reference low-dimensional embedding to target, as opposed to performing PCA or LDA on the input reference data.
- ncomponentsReference
Number of principal components for embedding reference data, given either as an integer or a named list for each reference dataset.
- ncomponentsSubset
Number of principal components for embedding query data prior to projecting to the reference, given either as an integer or a named list for each reference dataset.
- suppressMessages
Logical whether to suppress messages (default TRUE).
- projectAll
Logical whether to re-project reference data along with query (default FALSE).
- restrictFeatures
logical whether to restrict to features used in dimensionality reduction of reference data (default FALSE). Overall it's recommended that this be FALSE for single-hop integrations and TRUE for multi-hop integrations.
- maxFeatures
Maximum number of features to consider for predicting principal component scores (default 1000).
- plot
Logical whether to plot mosaic data UpSet plot and mosaic data topology networks (default TRUE).
- scale.center
Logical whether to re-center data to a mean of 0 (default FALSE).
- scale.scale
Logical whether to re-scale data to standard deviation of 1 (default FALSE).
Value
matrix containing common embedding with rows corresponding to cells, and columns corresponding to PCs or LDs for reference dataset(s).
Examples
set.seed(2021)
assay_list = mockMosaicData()
lapply(assay_list, dim)
#> $D1
#> [1] 150 50
#>
#> $D2
#> [1] 150 50
#>
#> $D3
#> [1] 150 50
#>
# specify which datasets to use as reference coordinates
reference_list = c("D1", "D3")
# specify some sample labels to distinguish using linear discriminant
# analysis (LDA)
labels_list = list(
D1 = rep(letters[1:5], length.out = ncol(assay_list[["D1"]]))
)
# examine the topology of this mosaic data integration
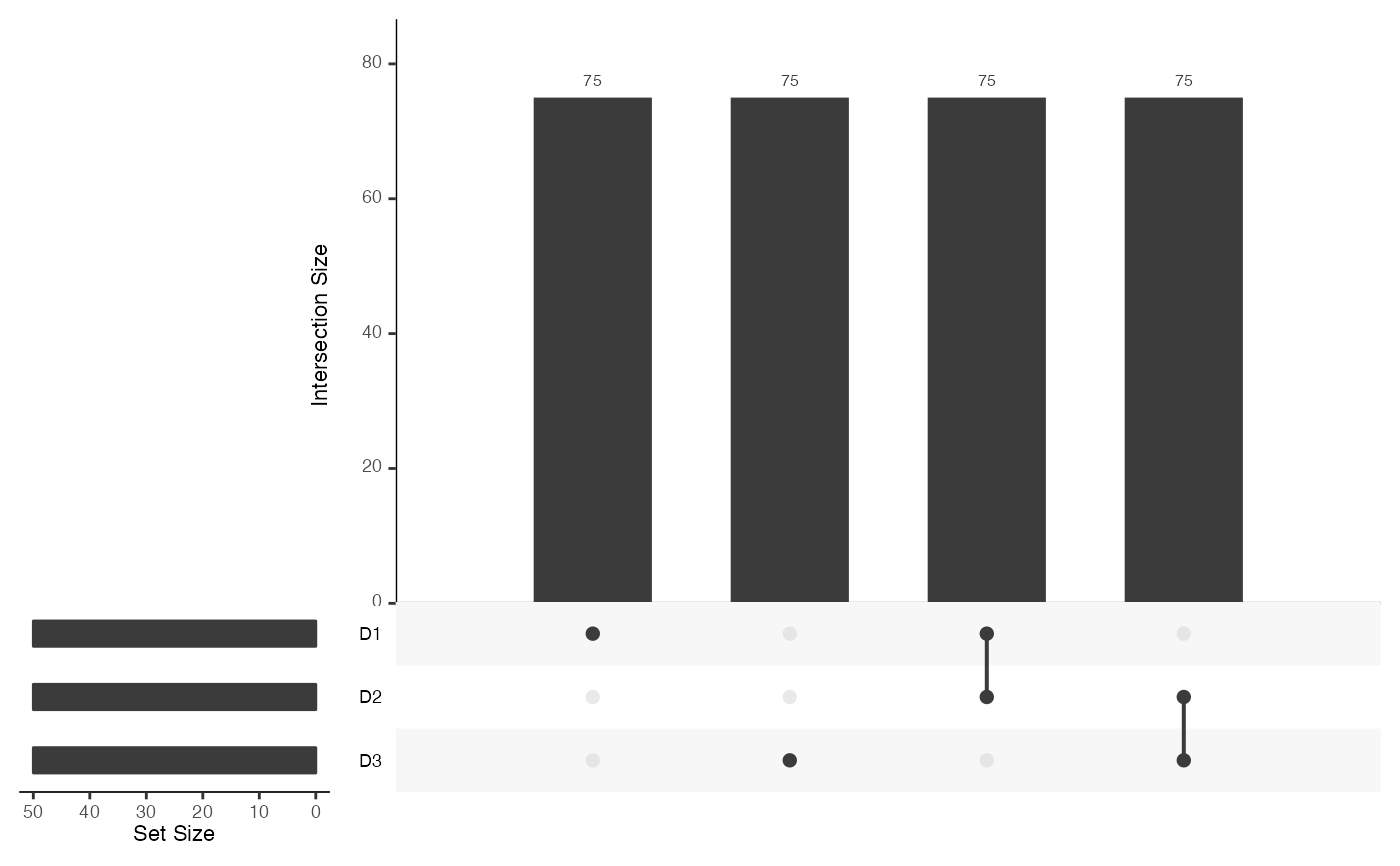
mosaicDataUpSet(assay_list)
#> Loading required package: UpSetR
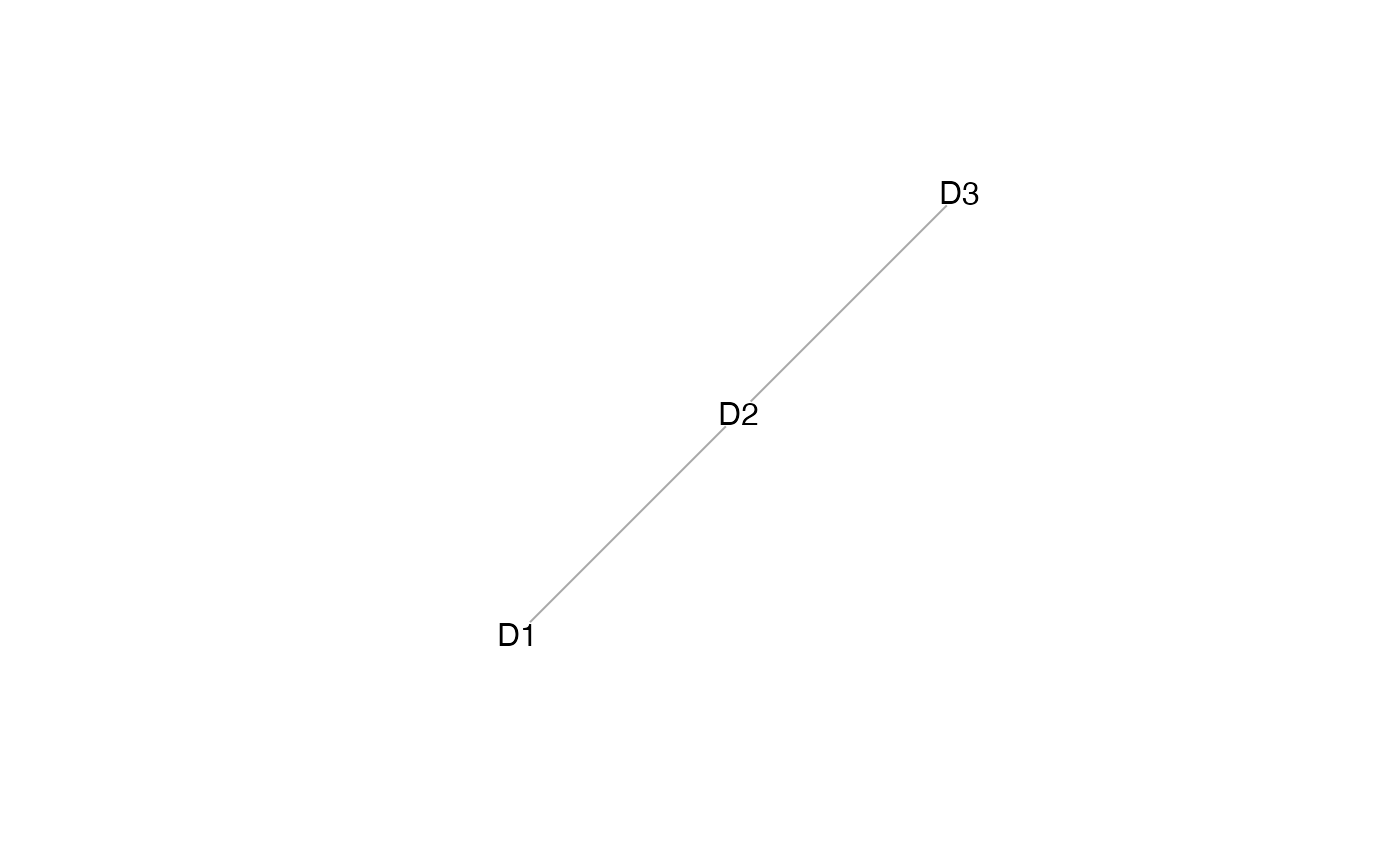
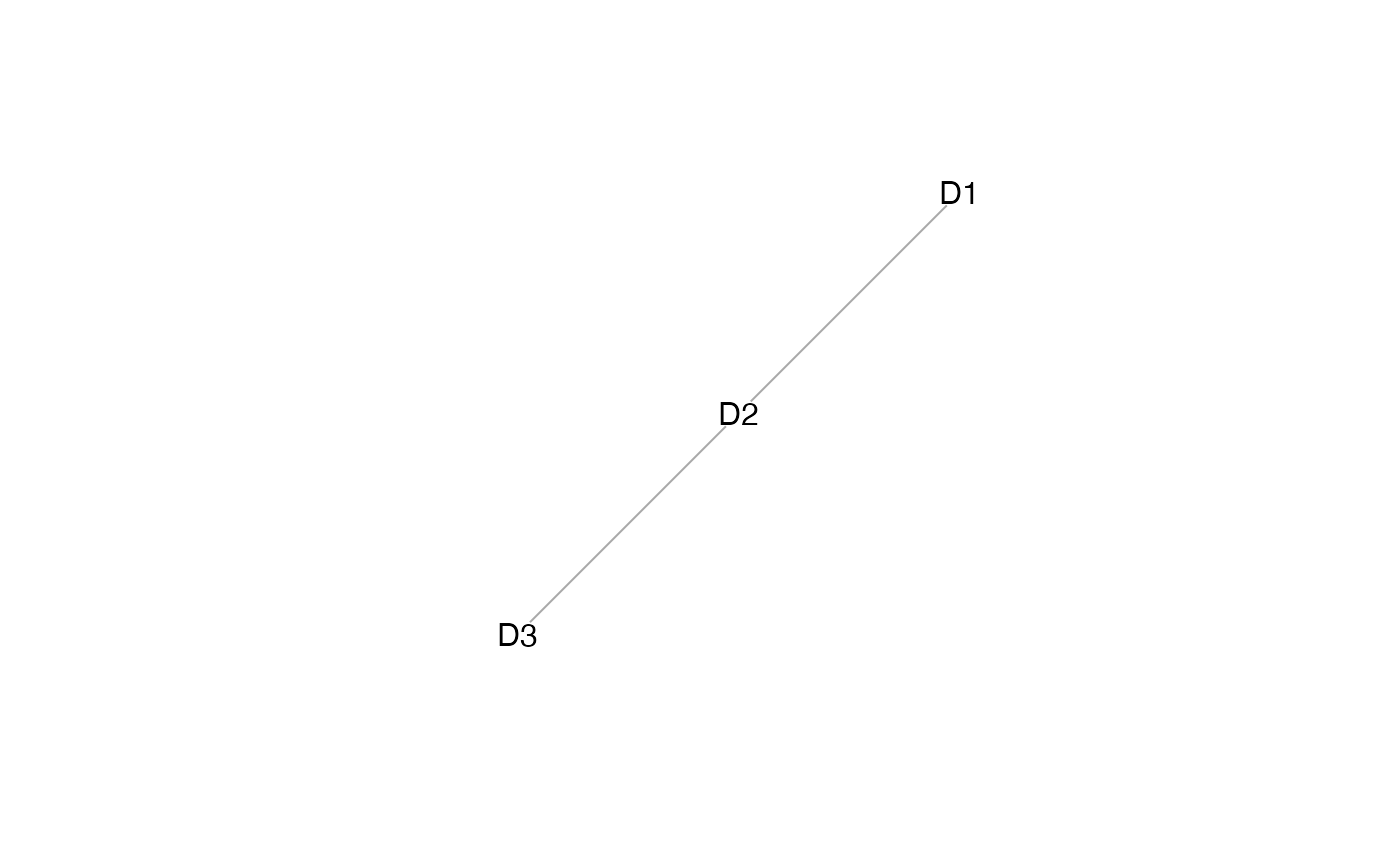
 plot(mosaicDataTopology(assay_list))
plot(mosaicDataTopology(assay_list))
 # stabMap
out = stabMap(assay_list,
reference_list = reference_list,
labels_list = labels_list,
ncomponentsReference = 20,
ncomponentsSubset = 20)
#> Loading required package: scater
#> Loading required package: SingleCellExperiment
#> Loading required package: SummarizedExperiment
#> Loading required package: MatrixGenerics
#> Loading required package: matrixStats
#>
#> Attaching package: ‘MatrixGenerics’
#> The following objects are masked from ‘package:matrixStats’:
#>
#> colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
#> colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
#> colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
#> colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
#> colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
#> colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
#> colWeightedMeans, colWeightedMedians, colWeightedSds,
#> colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
#> rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
#> rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
#> rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
#> rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
#> rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
#> rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
#> rowWeightedSds, rowWeightedVars
#> Loading required package: GenomicRanges
#> Loading required package: stats4
#> Loading required package: BiocGenerics
#>
#> Attaching package: ‘BiocGenerics’
#> The following objects are masked from ‘package:igraph’:
#>
#> normalize, path, union
#> The following objects are masked from ‘package:stats’:
#>
#> IQR, mad, sd, var, xtabs
#> The following objects are masked from ‘package:base’:
#>
#> Filter, Find, Map, Position, Reduce, anyDuplicated, append,
#> as.data.frame, basename, cbind, colnames, dirname, do.call,
#> duplicated, eval, evalq, get, grep, grepl, intersect, is.unsorted,
#> lapply, mapply, match, mget, order, paste, pmax, pmax.int, pmin,
#> pmin.int, rank, rbind, rownames, sapply, setdiff, sort, table,
#> tapply, union, unique, unsplit, which.max, which.min
#> Loading required package: S4Vectors
#>
#> Attaching package: ‘S4Vectors’
#> The following objects are masked from ‘package:base’:
#>
#> I, expand.grid, unname
#> Loading required package: IRanges
#> Loading required package: GenomeInfoDb
#> Loading required package: Biobase
#> Welcome to Bioconductor
#>
#> Vignettes contain introductory material; view with
#> 'browseVignettes()'. To cite Bioconductor, see
#> 'citation("Biobase")', and for packages 'citation("pkgname")'.
#>
#> Attaching package: ‘Biobase’
#> The following object is masked from ‘package:MatrixGenerics’:
#>
#> rowMedians
#> The following objects are masked from ‘package:matrixStats’:
#>
#> anyMissing, rowMedians
#> Loading required package: scuttle
#> Loading required package: ggplot2
#> treating "D1" as reference
#> generating embedding for path with reference "D1": "D1"
#> generating embedding for path with reference "D1": "D2" -> "D1"
#> generating embedding for path with reference "D1": "D3" -> "D2" -> "D1"
#> Loading required package: MASS
#> labels provided for "D1", adding LD components
#> Loading required package: Matrix
#>
#> Attaching package: ‘Matrix’
#> The following object is masked from ‘package:S4Vectors’:
#>
#> expand
#> generating embedding for path with reference "D1": "D1"
#> generating embedding for path with reference "D1": "D2" -> "D1"
#> generating embedding for path with reference "D1": "D3" -> "D2" -> "D1"
#> treating "D3" as reference
#> generating embedding for path with reference "D3": "D3"
#> generating embedding for path with reference "D3": "D2" -> "D3"
#> generating embedding for path with reference "D3": "D1" -> "D2" -> "D3"
# stabMap
out = stabMap(assay_list,
reference_list = reference_list,
labels_list = labels_list,
ncomponentsReference = 20,
ncomponentsSubset = 20)
#> Loading required package: scater
#> Loading required package: SingleCellExperiment
#> Loading required package: SummarizedExperiment
#> Loading required package: MatrixGenerics
#> Loading required package: matrixStats
#>
#> Attaching package: ‘MatrixGenerics’
#> The following objects are masked from ‘package:matrixStats’:
#>
#> colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
#> colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
#> colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
#> colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
#> colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
#> colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
#> colWeightedMeans, colWeightedMedians, colWeightedSds,
#> colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
#> rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
#> rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
#> rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
#> rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
#> rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
#> rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
#> rowWeightedSds, rowWeightedVars
#> Loading required package: GenomicRanges
#> Loading required package: stats4
#> Loading required package: BiocGenerics
#>
#> Attaching package: ‘BiocGenerics’
#> The following objects are masked from ‘package:igraph’:
#>
#> normalize, path, union
#> The following objects are masked from ‘package:stats’:
#>
#> IQR, mad, sd, var, xtabs
#> The following objects are masked from ‘package:base’:
#>
#> Filter, Find, Map, Position, Reduce, anyDuplicated, append,
#> as.data.frame, basename, cbind, colnames, dirname, do.call,
#> duplicated, eval, evalq, get, grep, grepl, intersect, is.unsorted,
#> lapply, mapply, match, mget, order, paste, pmax, pmax.int, pmin,
#> pmin.int, rank, rbind, rownames, sapply, setdiff, sort, table,
#> tapply, union, unique, unsplit, which.max, which.min
#> Loading required package: S4Vectors
#>
#> Attaching package: ‘S4Vectors’
#> The following objects are masked from ‘package:base’:
#>
#> I, expand.grid, unname
#> Loading required package: IRanges
#> Loading required package: GenomeInfoDb
#> Loading required package: Biobase
#> Welcome to Bioconductor
#>
#> Vignettes contain introductory material; view with
#> 'browseVignettes()'. To cite Bioconductor, see
#> 'citation("Biobase")', and for packages 'citation("pkgname")'.
#>
#> Attaching package: ‘Biobase’
#> The following object is masked from ‘package:MatrixGenerics’:
#>
#> rowMedians
#> The following objects are masked from ‘package:matrixStats’:
#>
#> anyMissing, rowMedians
#> Loading required package: scuttle
#> Loading required package: ggplot2
#> treating "D1" as reference
#> generating embedding for path with reference "D1": "D1"
#> generating embedding for path with reference "D1": "D2" -> "D1"
#> generating embedding for path with reference "D1": "D3" -> "D2" -> "D1"
#> Loading required package: MASS
#> labels provided for "D1", adding LD components
#> Loading required package: Matrix
#>
#> Attaching package: ‘Matrix’
#> The following object is masked from ‘package:S4Vectors’:
#>
#> expand
#> generating embedding for path with reference "D1": "D1"
#> generating embedding for path with reference "D1": "D2" -> "D1"
#> generating embedding for path with reference "D1": "D3" -> "D2" -> "D1"
#> treating "D3" as reference
#> generating embedding for path with reference "D3": "D3"
#> generating embedding for path with reference "D3": "D2" -> "D3"
#> generating embedding for path with reference "D3": "D1" -> "D2" -> "D3"
 head(out)
#> D1_PC1 D1_PC2 D1_PC3 D1_PC4 D1_PC5 D1_PC6
#> D1_cell_1 2.6270969 3.1709928 4.0506445 -3.609130459 -1.43056606 2.5013943
#> D1_cell_2 -4.5057485 -0.5752083 0.8340238 -1.014823114 -0.63151929 1.4937527
#> D1_cell_3 0.6226309 1.5472376 2.6103086 -0.018643114 2.81536539 1.0940201
#> D1_cell_4 -4.2263350 2.0444978 -0.5356968 -3.453648014 -2.92566847 -0.2716939
#> D1_cell_5 3.8947019 -0.9106738 -3.6039996 0.004893567 2.30739074 0.3341585
#> D1_cell_6 2.0177813 -3.5252245 -4.1753395 -1.148906355 -0.05251646 -1.8700432
#> D1_PC7 D1_PC8 D1_PC9 D1_PC10 D1_PC11 D1_PC12
#> D1_cell_1 -0.24764751 0.5873167 -2.3067376 3.2015090 -0.4526165 -1.2460048
#> D1_cell_2 -0.25444661 -0.7616007 -2.4968763 0.1378678 2.8105718 4.8446448
#> D1_cell_3 0.03710635 1.5760269 -0.3924504 1.1906449 -3.6871124 -3.7681881
#> D1_cell_4 -1.69233366 0.7617611 4.8401743 -2.0105921 -1.1875114 -1.5079628
#> D1_cell_5 -4.31581421 0.9061911 -0.9850855 1.2802993 -1.6064210 0.3283823
#> D1_cell_6 3.75335901 -0.4612008 -2.3790983 1.2309207 -2.1421033 -0.9491438
#> D1_PC13 D1_PC14 D1_PC15 D1_PC16 D1_PC17 D1_PC18
#> D1_cell_1 3.1375299 -4.378068 -3.444451 -1.78898621 -2.2372443 0.7444908
#> D1_cell_2 0.7939143 -2.721340 -0.377106 -0.09414872 1.4999731 -4.7338701
#> D1_cell_3 -0.4814744 3.724470 -1.594450 -1.49311501 1.9097362 -1.1429568
#> D1_cell_4 3.0741564 -1.146310 1.966764 2.49615083 -1.8948504 -0.2249584
#> D1_cell_5 0.2501072 1.337008 1.229278 -2.45020979 0.6301791 -1.0275208
#> D1_cell_6 -1.9202294 1.207617 2.005668 -0.81970304 -1.8491049 0.7083563
#> D1_PC19 D1_PC20 D1_LD1 D1_LD2 D1_LD3 D1_LD4
#> D1_cell_1 -1.2420356 -2.6537241 -0.3927678 -1.674139 -1.1392196 0.01773841
#> D1_cell_2 -1.5838926 -0.7553307 1.2724435 0.793653 1.1154764 1.00840563
#> D1_cell_3 2.0095487 -1.4511702 -1.4536953 1.599853 -0.9361014 0.06013095
#> D1_cell_4 0.2146626 1.1997077 1.1922330 -1.336241 -0.0218638 0.50445480
#> D1_cell_5 -2.5546764 -2.1599807 -1.8890462 -1.191700 -0.1356699 0.66198248
#> D1_cell_6 1.4360245 -0.1285014 0.1478705 -1.261443 0.7427569 -0.93665942
#> D3_PC1 D3_PC2 D3_PC3 D3_PC4 D3_PC5 D3_PC6
#> D1_cell_1 5.092261 8.2508095 3.425652 0.4690347 1.364696 -2.321035
#> D1_cell_2 12.595954 0.2080145 -14.737667 6.5019653 2.625458 -5.032633
#> D1_cell_3 -11.306183 -1.9951360 19.052222 -7.5799141 -4.386633 7.594893
#> D1_cell_4 -23.365322 -2.8256679 6.699823 -3.8222231 -12.100091 2.068094
#> D1_cell_5 -3.486373 1.6333765 12.325701 -6.0603755 6.831653 1.613624
#> D1_cell_6 -4.529260 2.8107633 5.415550 12.4524329 -4.605054 3.963515
#> D3_PC7 D3_PC8 D3_PC9 D3_PC10 D3_PC11 D3_PC12
#> D1_cell_1 3.8764340 4.0883439 2.428021 -0.7772289 -2.710610 -2.2746468
#> D1_cell_2 3.6140777 6.0370403 -9.409367 -7.6498222 7.823002 -0.9906758
#> D1_cell_3 -2.5210342 -4.6332347 9.397575 12.0328986 -10.564131 2.8742841
#> D1_cell_4 -0.1166194 0.9251444 12.568800 9.0282004 -1.519511 -0.4224119
#> D1_cell_5 -7.9823773 -5.9523524 9.895687 3.1960450 -12.193279 0.4332627
#> D1_cell_6 9.7299558 3.6624210 -2.562558 -2.4382349 2.061842 3.2577990
#> D3_PC13 D3_PC14 D3_PC15 D3_PC16 D3_PC17 D3_PC18
#> D1_cell_1 0.8287263 -2.576192 -1.231400 -2.2388540 -1.976509 5.664110
#> D1_cell_2 -6.5423054 7.724876 4.553024 14.3713652 8.397916 -2.623389
#> D1_cell_3 7.2103276 -13.081510 -4.107556 -17.0461773 -10.751769 1.107053
#> D1_cell_4 9.1727966 3.286651 -11.690377 -4.8886036 6.230477 2.364460
#> D1_cell_5 1.9820477 -14.167346 1.655577 -9.5230822 -12.926575 2.182262
#> D1_cell_6 3.3057180 14.400471 -8.922536 0.4140916 -4.918429 -2.660315
#> D3_PC19 D3_PC20
#> D1_cell_1 1.6945880 2.072783
#> D1_cell_2 -4.6158786 2.636875
#> D1_cell_3 7.0317033 -5.098531
#> D1_cell_4 -0.6198571 14.316522
#> D1_cell_5 7.6733032 -10.683882
#> D1_cell_6 -2.0122076 5.141252
head(out)
#> D1_PC1 D1_PC2 D1_PC3 D1_PC4 D1_PC5 D1_PC6
#> D1_cell_1 2.6270969 3.1709928 4.0506445 -3.609130459 -1.43056606 2.5013943
#> D1_cell_2 -4.5057485 -0.5752083 0.8340238 -1.014823114 -0.63151929 1.4937527
#> D1_cell_3 0.6226309 1.5472376 2.6103086 -0.018643114 2.81536539 1.0940201
#> D1_cell_4 -4.2263350 2.0444978 -0.5356968 -3.453648014 -2.92566847 -0.2716939
#> D1_cell_5 3.8947019 -0.9106738 -3.6039996 0.004893567 2.30739074 0.3341585
#> D1_cell_6 2.0177813 -3.5252245 -4.1753395 -1.148906355 -0.05251646 -1.8700432
#> D1_PC7 D1_PC8 D1_PC9 D1_PC10 D1_PC11 D1_PC12
#> D1_cell_1 -0.24764751 0.5873167 -2.3067376 3.2015090 -0.4526165 -1.2460048
#> D1_cell_2 -0.25444661 -0.7616007 -2.4968763 0.1378678 2.8105718 4.8446448
#> D1_cell_3 0.03710635 1.5760269 -0.3924504 1.1906449 -3.6871124 -3.7681881
#> D1_cell_4 -1.69233366 0.7617611 4.8401743 -2.0105921 -1.1875114 -1.5079628
#> D1_cell_5 -4.31581421 0.9061911 -0.9850855 1.2802993 -1.6064210 0.3283823
#> D1_cell_6 3.75335901 -0.4612008 -2.3790983 1.2309207 -2.1421033 -0.9491438
#> D1_PC13 D1_PC14 D1_PC15 D1_PC16 D1_PC17 D1_PC18
#> D1_cell_1 3.1375299 -4.378068 -3.444451 -1.78898621 -2.2372443 0.7444908
#> D1_cell_2 0.7939143 -2.721340 -0.377106 -0.09414872 1.4999731 -4.7338701
#> D1_cell_3 -0.4814744 3.724470 -1.594450 -1.49311501 1.9097362 -1.1429568
#> D1_cell_4 3.0741564 -1.146310 1.966764 2.49615083 -1.8948504 -0.2249584
#> D1_cell_5 0.2501072 1.337008 1.229278 -2.45020979 0.6301791 -1.0275208
#> D1_cell_6 -1.9202294 1.207617 2.005668 -0.81970304 -1.8491049 0.7083563
#> D1_PC19 D1_PC20 D1_LD1 D1_LD2 D1_LD3 D1_LD4
#> D1_cell_1 -1.2420356 -2.6537241 -0.3927678 -1.674139 -1.1392196 0.01773841
#> D1_cell_2 -1.5838926 -0.7553307 1.2724435 0.793653 1.1154764 1.00840563
#> D1_cell_3 2.0095487 -1.4511702 -1.4536953 1.599853 -0.9361014 0.06013095
#> D1_cell_4 0.2146626 1.1997077 1.1922330 -1.336241 -0.0218638 0.50445480
#> D1_cell_5 -2.5546764 -2.1599807 -1.8890462 -1.191700 -0.1356699 0.66198248
#> D1_cell_6 1.4360245 -0.1285014 0.1478705 -1.261443 0.7427569 -0.93665942
#> D3_PC1 D3_PC2 D3_PC3 D3_PC4 D3_PC5 D3_PC6
#> D1_cell_1 5.092261 8.2508095 3.425652 0.4690347 1.364696 -2.321035
#> D1_cell_2 12.595954 0.2080145 -14.737667 6.5019653 2.625458 -5.032633
#> D1_cell_3 -11.306183 -1.9951360 19.052222 -7.5799141 -4.386633 7.594893
#> D1_cell_4 -23.365322 -2.8256679 6.699823 -3.8222231 -12.100091 2.068094
#> D1_cell_5 -3.486373 1.6333765 12.325701 -6.0603755 6.831653 1.613624
#> D1_cell_6 -4.529260 2.8107633 5.415550 12.4524329 -4.605054 3.963515
#> D3_PC7 D3_PC8 D3_PC9 D3_PC10 D3_PC11 D3_PC12
#> D1_cell_1 3.8764340 4.0883439 2.428021 -0.7772289 -2.710610 -2.2746468
#> D1_cell_2 3.6140777 6.0370403 -9.409367 -7.6498222 7.823002 -0.9906758
#> D1_cell_3 -2.5210342 -4.6332347 9.397575 12.0328986 -10.564131 2.8742841
#> D1_cell_4 -0.1166194 0.9251444 12.568800 9.0282004 -1.519511 -0.4224119
#> D1_cell_5 -7.9823773 -5.9523524 9.895687 3.1960450 -12.193279 0.4332627
#> D1_cell_6 9.7299558 3.6624210 -2.562558 -2.4382349 2.061842 3.2577990
#> D3_PC13 D3_PC14 D3_PC15 D3_PC16 D3_PC17 D3_PC18
#> D1_cell_1 0.8287263 -2.576192 -1.231400 -2.2388540 -1.976509 5.664110
#> D1_cell_2 -6.5423054 7.724876 4.553024 14.3713652 8.397916 -2.623389
#> D1_cell_3 7.2103276 -13.081510 -4.107556 -17.0461773 -10.751769 1.107053
#> D1_cell_4 9.1727966 3.286651 -11.690377 -4.8886036 6.230477 2.364460
#> D1_cell_5 1.9820477 -14.167346 1.655577 -9.5230822 -12.926575 2.182262
#> D1_cell_6 3.3057180 14.400471 -8.922536 0.4140916 -4.918429 -2.660315
#> D3_PC19 D3_PC20
#> D1_cell_1 1.6945880 2.072783
#> D1_cell_2 -4.6158786 2.636875
#> D1_cell_3 7.0317033 -5.098531
#> D1_cell_4 -0.6198571 14.316522
#> D1_cell_5 7.6733032 -10.683882
#> D1_cell_6 -2.0122076 5.141252